How to Read a Decision Tree in R
Decision trees are a highly useful visual assistance in analyzing a series of predicted outcomes for a particular model. As such, it is often used every bit a supplement (or fifty-fifty culling to) regression analysis in determining how a serial of explanatory variables will impact the dependent variable.
In this particular example, we analyse the impact of explanatory variables of age, gender, miles, debt, and income on the dependent variable car sales.
Classification Problems and Decision Trees
Firstly, we load our dataset and create a response variable (which is used for the classification tree since we demand to catechumen sales from a numerical to categorical variable):

We then create the training and test data (i.e. the information that nosotros will employ to create our model and so the data we volition examination this data confronting):
#Create training and test data inputData <- fullData[1:770, ] # training data testData <- fullData[771:963, ] # examination data
Then, our classification tree is created:
#Classification Tree library(rpart) formula=response~Age+Gender+Miles+Debt+Income dtree=rpart(formula,information=inputData,method="grade",control=rpart.control(minsplit=30,cp=0.001)) plot(dtree) text(dtree) summary(dtree) printcp(dtree) plotcp(dtree) printcp(dtree)
Note that the cp value is what indicates our desired tree size – nosotros see that our Ten-val relative error is minimized when our size of tree value is 4. Therefore, the determination tree is created using the dtree variable by taking into business relationship this variable.
summary(dtree) Call: rpart(formula = formula, data = inputData, method = "class", command = rpart.control(minsplit = xxx, cp = 0.001)) north= 770 CP nsplit ane 0.496598639 0 2 0.013605442 1 3 0.008503401 vi 4 0.001000000 ten rel error xerror i 1.0000000 1.0000000 two 0.5034014 0.5170068 three 0.4353741 0.5646259 4 0.4013605 0.5442177 xstd ane 0.07418908 2 0.05630200 three 0.05854027 iv 0.05759793
Tree Pruning
The decision tree is and so "pruned", where inappropriate nodes are removed from the tree to preclude overfitting of the information:
#Clip the Tree and Plot pdtree<- prune(dtree, cp=dtree$cptable[which.min(dtree$cptable[,"xerror"]),"CP"]) plot(pdtree, uniform=TRUE, main="Pruned Nomenclature Tree For Sales") text(pdtree, employ.n=TRUE, all=True, cex=.viii)

The model is now tested confronting the examination information, and we see that we accept a misclassification percentage of sixteen.75%. Clearly, the lower the better, since this indicates our model is more accurate at predicting the "real" data:
#Model Testing out tabular array(out[1:193],testData$response) response_predicted response_input mean(response_input != response_predicted) # misclassification % [one] 0.2844156
Solving Regression Problems With Determination Trees
When the dependent variable is numerical rather than categorical, we will want to prepare a regression tree instead as follows:
#Regression Tree fitreg <- rpart(CarSales~Age+Gender+Miles+Debt+Income, method="anova", data=inputData) printcp(fitreg) plotcp(fitreg) summary(fitreg) par(mfrow=c(1,two)) rsq.rpart(fitreg) # cross-validation results
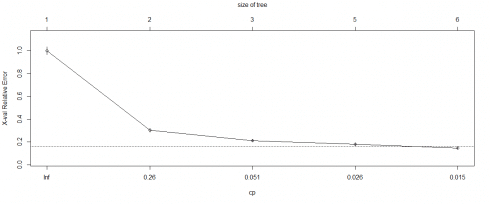
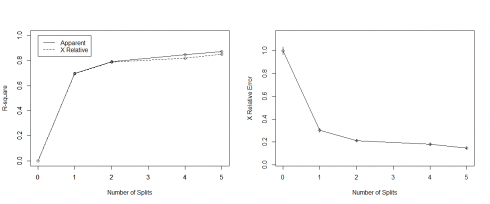
#Regression Tree fitreg printcp(fitreg) Regression tree: rpart(formula = CarSales ~ Age + Gender + Miles + Debt + Income, data = inputData, method = "anova") Variables actually used in tree structure: [1] Age Debt Income Root node mistake: 6.283e+10/770 = 81597576 northward= 770 CP nsplit rel mistake 1 0.698021 0 1.00000 ii 0.094038 1 0.30198 iii 0.028161 2 0.20794 4 0.023332 4 0.15162 5 0.010000 v 0.12829 xerror xstd i one.00162 0.033055 2 0.30373 0.016490 3 0.21261 0.012890 4 0.18149 0.013298 5 0.14781 0.013068
plotcp(fitreg) summary(fitreg) Call: rpart(formula = CarSales ~ Historic period + Gender + Miles + Debt + Income, data = inputData, method = "anova") n= 770 CP nsplit rel error 1 0.69802077 0 1.0000000 2 0.09403824 ane 0.3019792 3 0.02816107 2 0.2079410 iv 0.02333197 4 0.1516189 5 0.01000000 five 0.1282869 xerror xstd 1 ane.0016159 0.03305536 2 0.3037301 0.01649002 iii 0.2126110 0.01289041 4 0.1814939 0.01329778 5 0.1478078 0.01306756 Variable importance Debt Miles Income Age 53 23 20 4
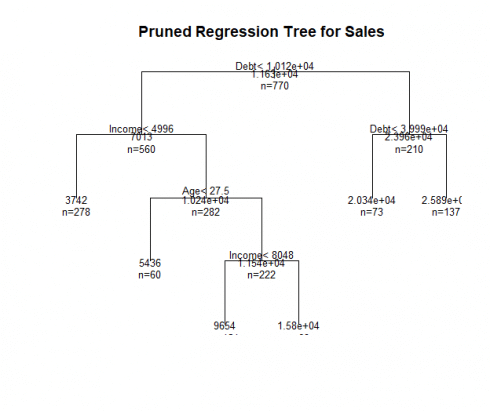
Now, we prune our regression tree:
#Prune the Tree pfitreg<- clip(fitreg, cp=fitreg$cptable[which.min(fitreg$cptable[,"xerror"]),"CP"]) # from cptable plot(pfitreg, uniform=TRUE, master="Pruned Regression Tree for Sales") text(pfitreg, apply.n=True, all=TRUE, cex=.eight)
Random Forests
However, what if we have many decision trees that nosotros wish to fit without preventing overfitting? A solution to this is to use a random forest.
A random forest allows us to determine the most important predictors beyond the explanatory variables past generating many decision trees and then ranking the variables by importance.
library(randomForest) fitregforest impress(fitregforest) # view results Telephone call: randomForest(formula = CarSales ~ Historic period + Gender + Miles + Debt + Income, information = inputData) Type of random woods: regression Number of trees: 500 No. of variables tried at each split: 1 Hateful of squared residuals: 10341022 % Var explained: 87.33 > importance(fitregforest) # importance of each predictor IncNodePurity Age 5920357954 Gender 187391341 Miles 10811341575 Debt 21813952812 Income 12694331712
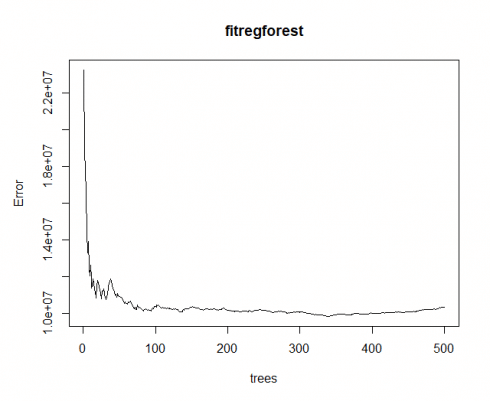
From the above, we see that debt is ranked equally the most important factor, i.e. customers with high debt levels will be more likely to spend a greater amount on a car. Nosotros see that 87.33% of the variation is "explained" past our random wood, and our error is minimized at roughly 100 trees.
Source: https://datascienceplus.com/decision-trees-and-random-forests-in-r/
0 Response to "How to Read a Decision Tree in R"
Enviar um comentário